Innovation in Bioanalytical Research for Translational Medicine: This Time It’s Personal
Modern medicine has usually taken a “one size fits all approach” to therapies. Pharmaceutical companies develop drugs for the populations instead of an individual. 
The “one size fits all approach” is driven by economics. Developing a new drug can cost over a billion dollars. It can be ten or more years before the drug reaches patients. Pharmaceutical companies develop drugs for the largest possible population to recoup costs.
Patients tend to suffer in this impersonal approach. They see themselves as anonymous, voiceless subjects at the whims of the pharmaceutical industry. They may feel that their conditions are not important enough to merit therapeutic development.
There are programs to develop drugs for rare diseases. Drugs and biologics intended to treat fewer than 200,000 people in the USA are given “Orphan status”, according to the Orphan Drug Act of 1983. This status helps companies recoup the higher development and marketing costs of these drugs. Over 600 drugs have come into the market as a result of “Orphan Status.”
Yet there are conditions that affect very small populations. There may never be drugs developed to treat extremely rare conditions. The good news is that technology is improving on multiple fronts. Some of these gains will result in more personalized medicine. In addition, the 21st Century Cures act emphasizes the “voice of the patient.”
Following is a discussion on a few technologies and methodologies to personalize medicine.
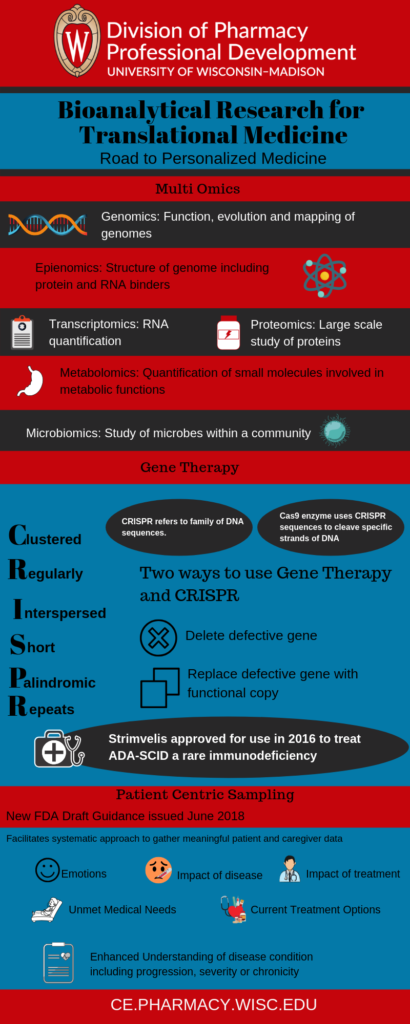
Multi-omics
High-throughput genotyping, large data sets and statistical tools have helped scientists. create a quality reference map of the human genome. As a result, thousands of gene variants (rare and common) contributing to disease have been mapped.
However, the identified loci thus far are responsible for only a small part of the heritable component for specific diseases. Also, Mendelian diseases typically result from changes in coding regions of genes while many common diseases are usually the result of changes in gene regulation. The environment and genetic background can also influence the health of those with the identified loci.
Thus, genetic data alone does not give a complete picture of disease susceptibility. It is necessary to integrate other data sets to better identify molecular patterns associated with a disease. “Omics” is a suffix added to a specific field of study in biology. It implies a comprehensive or global assessment of a set of molecules.
Today there are over a dozen different omics fields. The following is a list of most well-known omics fields.
- Genomics was the first discipline to appear. This field has rapidly expanded. Technological advances have created cost-efficient, high throughput analysis of biologic molecules.
- Epigenomics followed genomics. It examines the structure of the genome. This includes protein and RNA binders, alternative DNA structures.
- Transcriptomics studies RNA quantification genome-wide
- Proteomics is a large-scale study of proteins. It includes peptide abundance, modification and interaction
- Metabolomics is the quantification of multiple small molecule types involved in metabolic functions
- Microbiomics studies microorganisms within a community. The gut microbiome is a popular subject in this area. Other communities include mucosal membranes and the skin.
Each of these omics datasets can provide their own list of differences associated with various diseases. These datasets can serve as markers of the disease process. They also offer insight into the biological pathways or processes involved in healthy or disease states. Individually, each dataset is limited to correlations. These correlations typically reflect reactive processes rather than causative ones. Integrating the different omics datasets provides a holistic overview. This information can help identify potential disease causative changes or targets for treatment.
Complex diseases have many causes. Combinations of a variety of factors could be causing the same disease. These diseases typically develop over time. They may involve both genetic and environmental factors. A comprehensive insight will require coordinated sets of multiple omics data. This data must be collected at multiple time points and from multiple relevant tissues. The utility of multi-omics will increase as large and diverse data sets are created.
In the future, it may be possible to track individual health indicators. Doctors may be able to prescribe a personalized plan of preventative measures. They may also be able to use the individualized data to identify the most effective therapies.
 Gene Therapy
Gene Therapy
Scientists have used the human genome map to identify many disease-causing genes. Some diseases are caused by a single gene. Most diseases, though, are caused by multiple genes as well as environmental factors.
Gene therapy attempts to cure the patient by fixing defective genes. This can be done with potentially no side effects. It is ideal for diseases caused by a single or a few functional genes. There are some well-known diseases that may be good candidates for gene therapy. These include:
- Parkinson’s Disease
- Huntington’s Disease
- Sickle Cell Anemia
- Inflammatory arthritis
- Some cancers and others.
There are two approaches to cure disease using gene therapy.
- Deleting a gene that was responsible for a defective protein
- Replacing a defective gene with a functional copy
In both cases, the challenge is delivering the therapy to the affected cells. There are still some unanswered questions around gene therapy. Once the target is hit will only one treatment solve the problem? Will multiple doses and follow-up be required?
The most famous gene therapy tool is CRISPR. CRISPR is an acronym for Clustered Regularly Interspaced Short Palindromic Repeats. These are a family of DNA sequences found in prokaryotic organisms. These sequences are derived from DNA fragments of viruses that previously infected the organism. They are used to detect and destroy DNA from similar viruses during subsequent infections.
Cas9 (CRISPR-associated 9) is an enzyme that uses CRISPR sequences to recognize and cleave specific strands of DNA. The CRISPR/Cas9 technology can be used to edit genes and has the potential to treat diseases. Like other gene therapies, CRISPR is still in the experimental stages. Currently, there are no FDA approved gene therapies regardless of mechanism.
However, there is one gene therapy that is approved for use in Europe. Strimvelis was approved in 2016 to treat a rare disease called ADA-SCID (Severe Combined Immunodeficiency due to Adenosine Deaminase Deficiency). It is estimated to occur in approximately 15 patients per year in Europe (and 12 in the US where the therapy is not approved).
The current treatment standard is weekly enzyme replacement therapy. Clinical trials of Strimvelis were impressive with a 100% survival rate. At a seven-year median follow up 75% of the patients no longer needed enzyme replacement therapy. However, the treatment is expensive. It is twice the annual cost of enzyme replacement. However, only one treatment is necessary vs. weekly enzyme replacement injections. To date only five patients have used it. GSK, the developer of this product, sold it to Orchard Therapeutics in March 2018.
Gene therapy may still be in the early stages of development. However, in the not too distant future, it is likely that more therapies like Strimvelis will reach patients who need them.
One crucial element that needs to be addressed is funding. Developing therapies for patient populations in the hundreds is not economically viable. It is unlikely that GSK recouped its development expenses for Strimvelis. Will the potentially low or negative returns hinder development? This issue must be resolved as technology makes individualized therapies possible.
Patient-Centric Sampling
The FDA issued new draft guidance in June 2018 focusing on sampling methods for patient experiences. The draft lays out broad suggestions for the kinds of patient experiences that might be relevant. These suggestions include:
- The patient’s own feelings about potential or current treatments
- Impact of disease on the patient
- Impact of treatment on the patient
- Unmet medical needs
- Currently available treatment options
- An enhanced understanding of the natural history of the disease or condition, including progression, severity, chronicity
This draft guidance was mandated under the 21st Century Cures Act. It intends to facilitate the advancement and use of systematic approaches to collect meaningful patient and caregiver data to help better inform medical product development.
“Over the last several years we’ve seen many biopharma companies create programs intended to involve the patient voice in their development programs, and the increasing strength of patient advocacy groups engaging in drug development, but patient input has been largely regarded as a ‘nice to have.’ This guidance represents a concrete step in the FDA saying that they intend patient input to be part of regulatory decision making. Sponsors of medical product development programs are going to have to think about the collection of information and data from patients in a more structured and formal way than most have been doing so far, and this guidance provides information about how to do that.”
Linda McNair, CMO at WIRB-Copernicus Group
This draft guidance also encourages the use of social media as a way of gauging patient attitudes, something the FDA has not done previously.
“We learn through scientific advances, but also by listening to patients. Our work demands that we must continue to reflect on how we can make the science of drug development and review more modern and more patient-centered, so that approved products impact the metrics that real-world patients and families value most. This requires ongoing engagement with the patient community.”
FDA Commissioner Scott Gottlieb in a statement released in support of the draft guidance.
After many years of being ignored in the drug development process, the patient is finally going to be heard. This is a welcome development to help empower the end user of all drug development projects. Those who work in drug development may forget that they, too, will be that patient at some point in their life. Having a say in treatments benefits us all.
Technological advances and paradigm shifts are giving the patient a voice in their treatment. It is also potentially creating better odds for success. Multi-omics, gene therapy and patient-centric sampling are part of this process. Successful pharmaceutical scientists need to know the latest updates in these fields.
20th Annual Land O’ Lakes Bioanalytical Conference
The 2019 Land O’Lakes Bioanalytical Conference will provide a forum for pharmaceutical scientists, researchers, and regulators to discuss the latest updates in these and other areas. The 2019 conference theme is “Innovation in Bioanalytical Research for Translational Medicine: New Modalities, New Technologies, and Evolving Regulations.” Presentations will focus on regulatory updates in gene therapy, companion diagnostics, patient-centric bioanalysis, advances in multi omics miniaturization, and more.
The conference will be held at the Fluno Center at the UW-Madison Campus from July 15-18, 2019.
The Land O’Lakes series is a premier conference series for pharmaceutical scientists. With over 60 years of experience, it is one of the oldest and most respected conference series. In addition to the Bioanalytical Conference, Land O’ Lakes series includes the Annual Pharmaceutical Analysis Conference (August 12-15, 2019) and DMPK Conference (September 9-12, 2019).
Discover all Land O’ Lakes Conferences
References
https://www.centerwatch.com/cwweekly/2018/06/18/fda-draft-guidance-on-patient-centric-drug-development-focuses-on-sampling-methods/
https://www.technologyreview.com/s/601390/gene-therapys-first-out-and-out-cure-is-here/
https://genomebiology.biomedcentral.com/articles/10.1186/s13059-017-1215-1
http://omics.org/
https://geewisc.wisc.edu/
