Endogenous Biomarkers: A tool to detect drug-drug interactions (DDIs)
DDI Risks
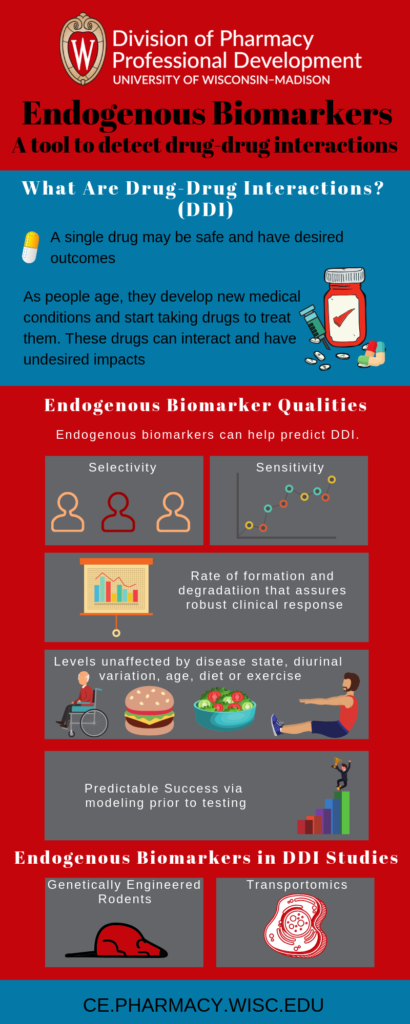
As the population ages, many patients will be taking more than one drug at a time. Multiple drugs can interact with each other and cause undesired side effects. These drug-drug interactions (DDIs) can make previously safe (if taken alone) drugs become dangerous. Thus, regulatory agencies require new drug applications to include clinical pharmacokinetic DDI studies. DDI studies evaluate the effect of drug candidates on co-administered drugs and vice-versa.
Interaction risks are usually evaluated before first in human (FIH) trials. This is done with in-vitro studies using cell-based systems. This approach has been well established for drug metabolizing enzymes in the CYP450 family. However, it isn’t as straightforward for drug transporters.
Regulatory agencies, including the FDA, have issued guidances on DDI studies. These guidances determine if DDI studies are necessary, based on in vitro transporter data. In vitro data are usually accurate in predicting patient DDI risk. But there are limitations in using in vitro inhibition data to predict transporter related DDIs. Regulatory guidances are conservative to reduce the number of false negatives. This has resulted in an increased number of required clinical DDI studies.
The limitations of in vitro transporter data may be resolved with endogenous biomarkers. Many endogenous biomarkers can be measured in plasma and urine. These measurements can serve as early indicators of potential transporter-mediated DDIs.
Recently several endogenous compounds have been identified as potential DDI biomarkers. These biomarkers are substrates for drug transporters in the kidney and liver. Therapeutic doses and exposure for efficacy are not established during early clinical trials. That is why sensitive biomarkers are critical to assessing DDI risk in this stage. For example – DDIs with statins for drugs developed to treat diabetes could stop a program in its tracks.
Transporters in the liver and kidney help modulate the hepatic and renal elimination of drugs. To understand the DDI potential of these drugs, it is crucial to predicting their inhibitory effect on these transporters. Endogenous biomarkers can help predict this inhibitory effect. This makes them important in DDI risk assessment.
 Predictive Endogenous Biomarkers
Predictive Endogenous Biomarkers
Extensive characterization can help ensure the suitability of predictive biomarkers. Due to limited data, the search for suitable transporter biomarkers is primarily empirical. Ideal biomarkers show the following traits
- Selectivity
- Sensitivity
- Rate of formation and degradation that assures a robust clinical response
- Levels unaffected by disease state, diurnal variation, age, diet or exercise
- Predicted success via modelling prior to testing
Various approaches have been used to identify biomarker candidates for transporters. Some selections were made by mining literature data. This data reports substrate specificity as determined by in vitro transport studies in recombinant systems.
It is not easy to know to what extent in vitro substrates will be relevant in vivo. It is also difficult to predict the ratio of transport by the transporter relative to the total uptake and efflux of an endogenous substrate. Specificity also needs to be established. Genome wide association studies (GWAS) can be helpful for pathway characterization. They demonstrate the relationship between polymorphisms and plasma or urine levels of desired biomarkers.
Genetically engineered rodent models are a vital tool in DDI studies. These models lacking specific transporters help determine the relevance of a transporter in affecting disposition of a biomarker. However, orthologues do not exist for all transporters. Relative abundance and substrate specificity of transporters differs across species. Thus, direct extrapolation from other species to humans is difficult.
Transportomics is a methodology developed to identify novel substrates for ABC transporters. Compounds are introduced to inside-out membrane vesicles. These vesicles contain the transporter of interest as well as control vesicles without them. Compounds that are detected in the transporter containing vesicles but not in the controls are considered substrates. Another method used to identify novel substrates is application of targeted metabolomics using knockout mice. These methods are still evolving and substrate identification is tedious.
22nd Annual DMPK Conference
Endogenous biomarkers are becoming a useful approach to assess the DDI liability of drug candidates during drug development. One of the plenary session at the 22nd Annual DMPK Conference is dedicated to endogenous biomarkers.
This year’s theme is “Found in Translation: Adaptive DMPK Strategies.” Other plenary session topics include:
- Target Mediated Drug Disposition (TMDD)
- Disposition in New and Non-traditional therapeutic mechanisms
- Regulatory updates
- Microphysiological systems
You will not want to miss this opportunity to keep up with DMPK and network with industry experts. Join us at the Fluno Center on the UW-Madison Campus from September 9-12, 2019. To learn more and register, go to:
The Land O’Lakes series is a premier conference series for pharmaceutical scientists. With over 60 years of experience, it is one of the oldest and most respected conference series. In addition to the DMPK Conference, the 2019 Land O’ Lakes series includes the Bioanalytical Conference (July 15-18, 2019) and Pharmaceutical Analysis Conference (August 12-15, 2019).
Discover all Land O’ Lakes Conferences
References
- Chu X, Hoyee Chan G, Evers R, Identification of Endogenous Biomarkers to Predict the Propensity of Drug Candidates to Cause Hepatic or Renal Transporter-Mediated Drug-Drug Interactions, Journal of Pharmaceutical Sciences 2017; 106(9): 2357-2367
- Yee SW, Giacomini MM, Hsueh CH, et al. Metabolomic and genome-wide association studies reveal potential endogenous biomarkers for OATP1B1.
- Clin Pharmacol Ther. 2016;100(5):524-536